
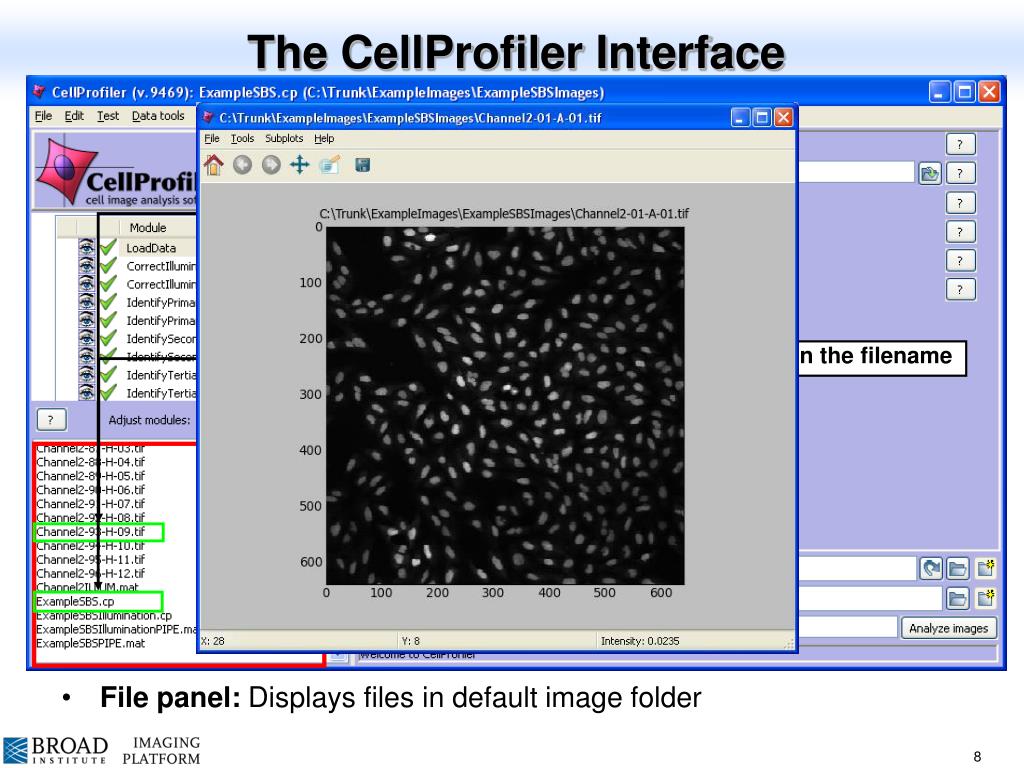
The main module that will need changed for the pipelines written by the facility will be module titled “IdentifyPrimaryObjects” so it’s a good idea to test this module at the very least. Advice may be written here regarding which parameters to change if the modules aren’t giving you the desired results.  For each module, there is a note section at the top. This means things really change depending on the camera and zoom you are using compared to what was used to set up the pipeline. One major difference with CellProfiler to other image analysis programs is that analysis is performed using pixels instead of microns or another measurement unit. If you want to how the pipeline performs on a different image click “Next Image Set”, this will bring you back to the top of the pipeline with the next image set. If you can’t find the resultant window you can double click on the module name in the pipeline to bring it to the front. A new window pops up for each module once it has run. To change each module between open and closed you can double click the icon there. To see the result the little eye icon to the left of the module needs to open. The module you are currently on (the active module) will be in bold and underline. By clicking the “Step” button you can see the results of each module. To test the pipeline on a couple of your images, click the “Start Test Mode” button in the bottom left. An example of each pipeline like this can be found in the repo called “MicroManagerExamplePipeline_split” and “MicroManagerExamplePipeline” respectively with images to try the pipeline on. This is due to CellProfiler reading ome-tiffs as multi-frame TIFFs instead of multi-channel. You could also change the filter settings to say “Metadata have frame matching” instead of “C matching”. An example of this is in our example pipelines folders. Name all your images thing in the next module and then use a module called “ColorToGrey” to split the channels. If you have Micro-Manager images, it’s easier to skip metadata and this step completely (even though they are multi-channel). The names are important as they are what are used to apply the analysis in the pipeline. If you images differ from this you will need to change which channel has which name. Most pipelines have assumed that your images are 3 channels and they are in wavelength order (i.e. You may remember this from capturing them but you can only also open them in another image analysis program to find out. To know if the channels in your images have the correct names then you will need to know the order of your images. You will see that there are “rules” for naming images, for example, that images with “Metadata with C matching 0” will be named DAPI. Next, check the NamesAndTypes modules to check what names are being assigned to each channel. After doing this, you should see the table populate with your images with a line per channel. First click the “Update Metadata” button in the top half of the module and then click the “Update” button on the table in the bottom half. If you have loaded multi-channel images then you will need to do the below:
For each module, there is a note section at the top. This means things really change depending on the camera and zoom you are using compared to what was used to set up the pipeline. One major difference with CellProfiler to other image analysis programs is that analysis is performed using pixels instead of microns or another measurement unit. If you want to how the pipeline performs on a different image click “Next Image Set”, this will bring you back to the top of the pipeline with the next image set. If you can’t find the resultant window you can double click on the module name in the pipeline to bring it to the front. A new window pops up for each module once it has run. To change each module between open and closed you can double click the icon there. To see the result the little eye icon to the left of the module needs to open. The module you are currently on (the active module) will be in bold and underline. By clicking the “Step” button you can see the results of each module. To test the pipeline on a couple of your images, click the “Start Test Mode” button in the bottom left. An example of each pipeline like this can be found in the repo called “MicroManagerExamplePipeline_split” and “MicroManagerExamplePipeline” respectively with images to try the pipeline on. This is due to CellProfiler reading ome-tiffs as multi-frame TIFFs instead of multi-channel. You could also change the filter settings to say “Metadata have frame matching” instead of “C matching”. An example of this is in our example pipelines folders. Name all your images thing in the next module and then use a module called “ColorToGrey” to split the channels. If you have Micro-Manager images, it’s easier to skip metadata and this step completely (even though they are multi-channel). The names are important as they are what are used to apply the analysis in the pipeline. If you images differ from this you will need to change which channel has which name. Most pipelines have assumed that your images are 3 channels and they are in wavelength order (i.e. You may remember this from capturing them but you can only also open them in another image analysis program to find out. To know if the channels in your images have the correct names then you will need to know the order of your images. You will see that there are “rules” for naming images, for example, that images with “Metadata with C matching 0” will be named DAPI. Next, check the NamesAndTypes modules to check what names are being assigned to each channel. After doing this, you should see the table populate with your images with a line per channel. First click the “Update Metadata” button in the top half of the module and then click the “Update” button on the table in the bottom half. If you have loaded multi-channel images then you will need to do the below: 
After you’ve added your images, go to the next module ‘Metadata’.CellProfiler can read any type of file format so there is no need to convert your files before use.
To do this, navigate to this module by double clicking on it and dragging you files into the big area that says “Drop Files and Folders here.” CellProfiler processes any image files found in any folders dragged in so don’t worry about putting all images in the same folder. Images is where you tell CellProfiler which images you want to be analysed.The first four modules which will be in any pipeline are ‘Images’, ‘Metadata’, ‘NamesAndTypes’ and ‘Groups’.Open CellProfiler and in the main window go to File > Import > Import from URL.
CELLPROFILER PIPELINES CODE
If you click the "raw" button near the top right of the top of the code you will go to a new page where the script code is shown in raw form.Go to the pipeline you want here in this repository (for example - ).Alternatively, you can go to ‘File’ > ‘Import’ > ‘Pipeline from file’.Once you drag it you will be asked if you want to load the pipeline to which you can say “Yes”. cpipe file over to the white window on left side of the GUI. The easiest way to load a CellProfiler pipeline is to drag the.Keep reading for some help on using the pipelines. This repository contains CellProfiler Pipelines written for users of the IGC AIR facility.







 0 kommentar(er)
0 kommentar(er)
